SeqCode: produceTESmaps
General description
Generation of density heat maps of ChIPseq/ATACseq/RNAseq samples around the TES of genes.
> bin/produceTESmaps -h SeqCode_v1.0 User commands produceTESmaps NAME produceTESmaps - a program to produce ChIPseq/ATACseq/RNAseq heat maps around the TES. SYNOPSIS produceTESmaps [-bBfFH <Rcolor>][-d][-i <real>][-l <bp>][-n][-w <bp>][-v][-x prefix][-h] <chrom_info> <refGene.txt> <SAM file> <targetGenes.txt> <name> <length> OUTPUT One folder with 5 files: - Numerical distribution of reads. - Numerical distribution of reads (processed). - Ranking of genes in the map. - R script commands. - Graphical representation (R is required). OPTIONS -b : Background or color1 (default: white). -B : General background color (default: white). -d : Demo mode for small BAM files (min number reads control off). -f : Foreground or color2 (default: black). -F : General foreground color (default: black). -H : Heatmap background color (default: white). -i : Increase the color intensity (default: 0). -l : Avg. fragment size (default: 150). -n : Normalize using the total number of reads. -s : Number of spike-in reads for reference-adjusted normalization. -t : Noise reduction cutoff (default: 1.5). -w : Window resolution (default: 100). -v : Verbose. Display info messages. -x : Prefix for the output folder. -h : Show this help. SEE ALSO SeqCode homepage: http://ldicrocelab.crg.es GitHub source code: https://github.com/eblancoga/seqcode AUTHORS Written by Enrique Blanco. SeqCode_v1.0 User commands produceTESmaps
Examples
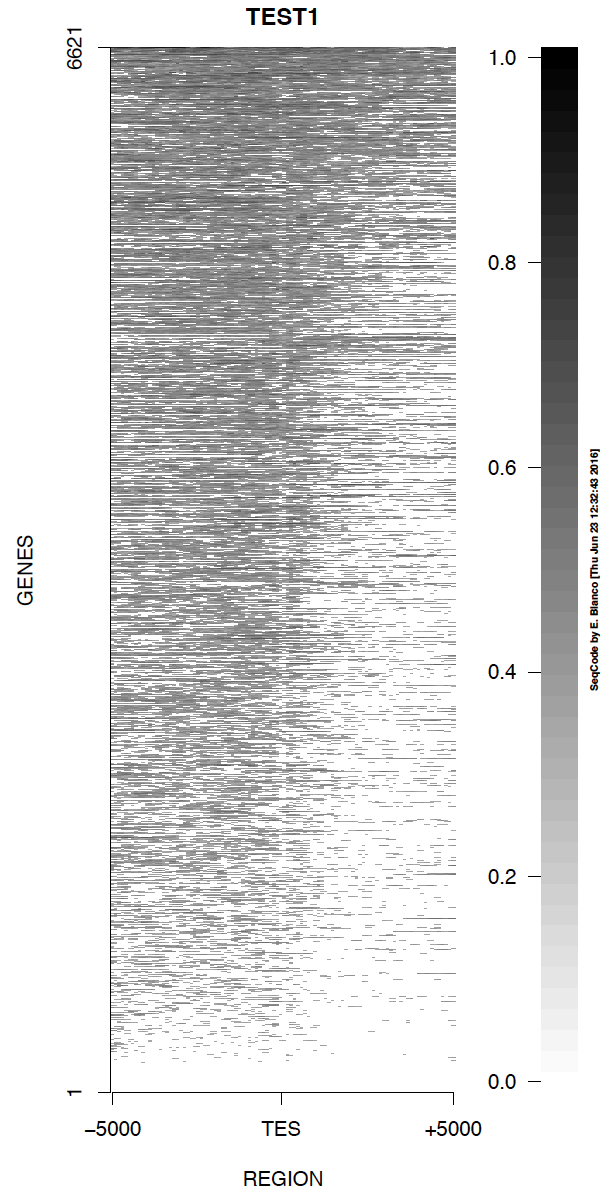
Example 1. How to generate a basic ChIPseq density heat map around the TES of genes.

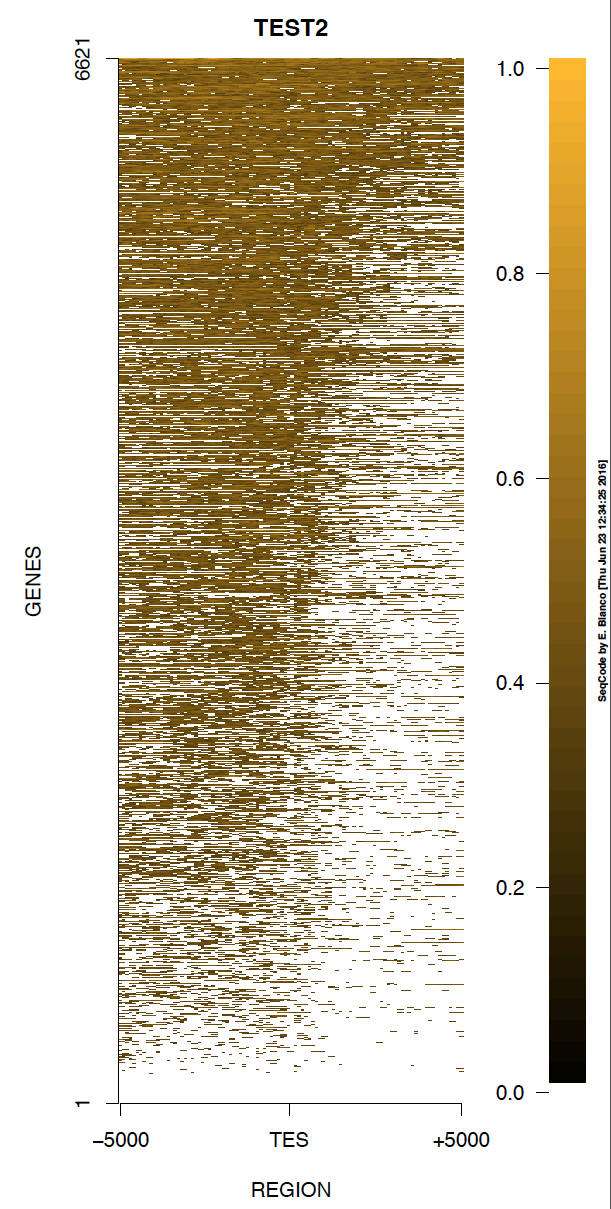
Example 2. How to change the range of colors into the heat map of genes (background is white).
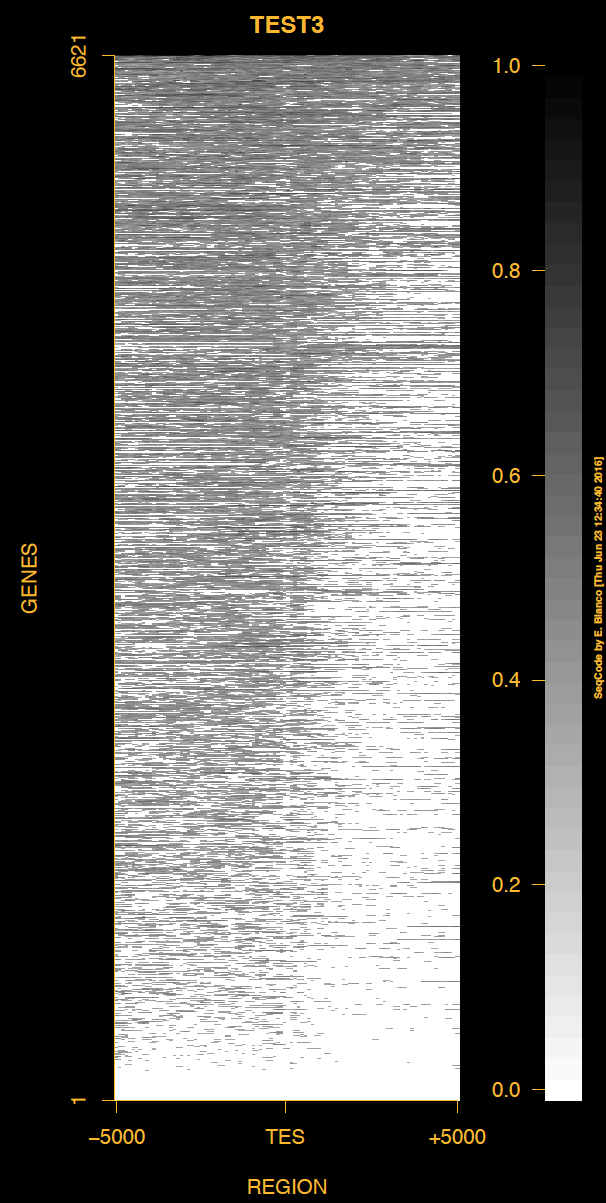
Example 3. How to change the outer background and foreground colors of the heat map of genes.

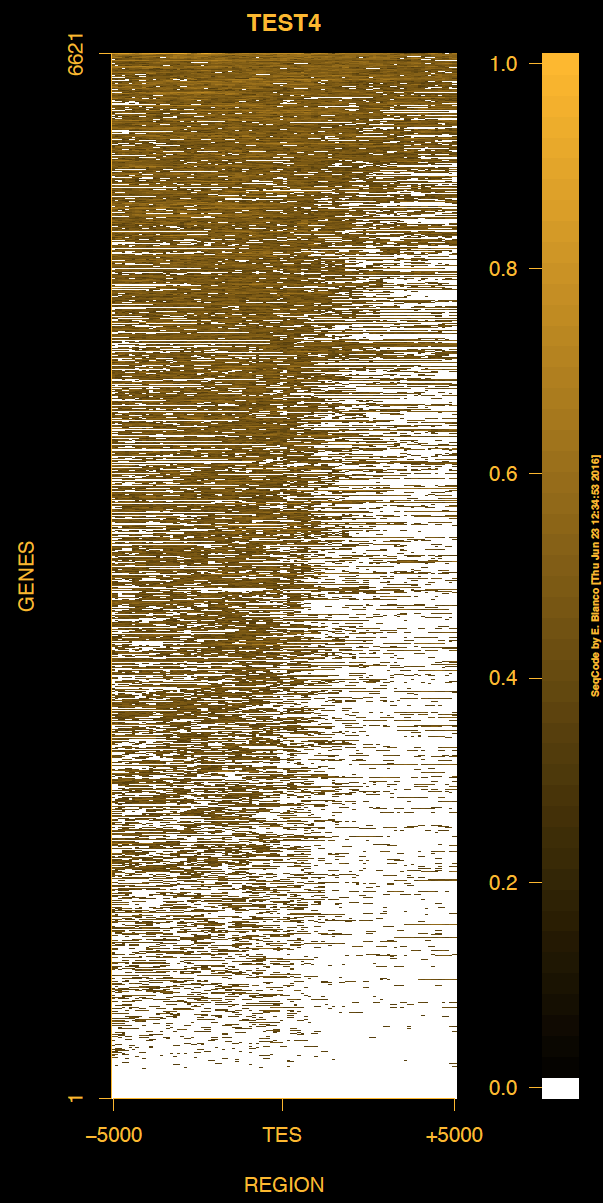
Example 4. How to change the outer background and foreground colors and the range of the heat map of genes (background is white).
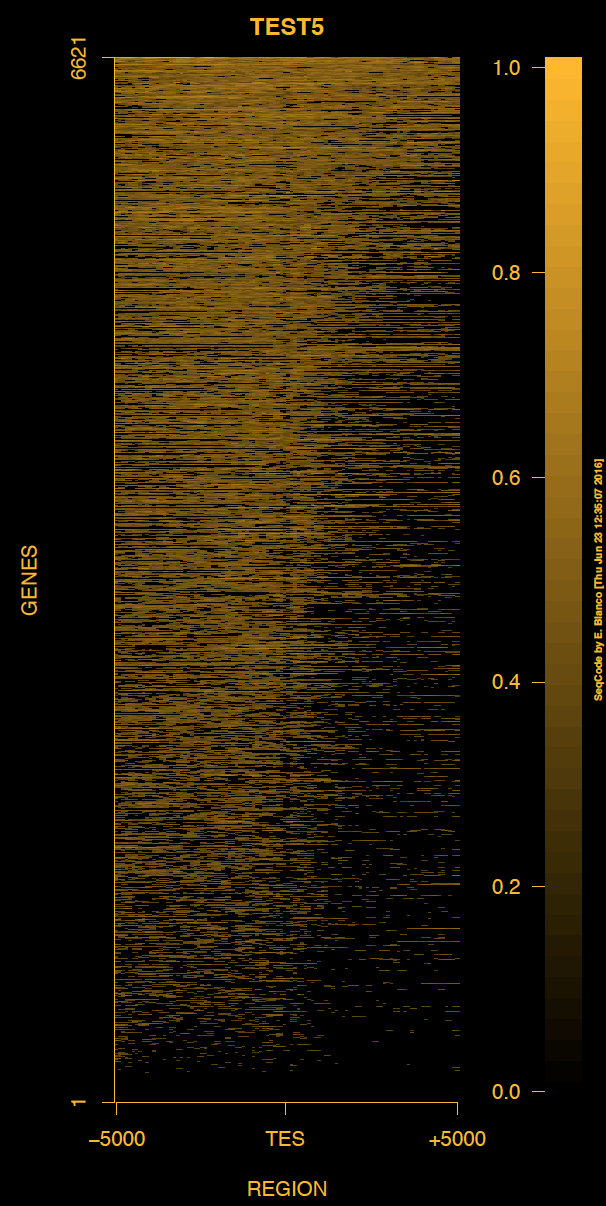
Example 5. How to change the full schema of colors of the heat map of genes.
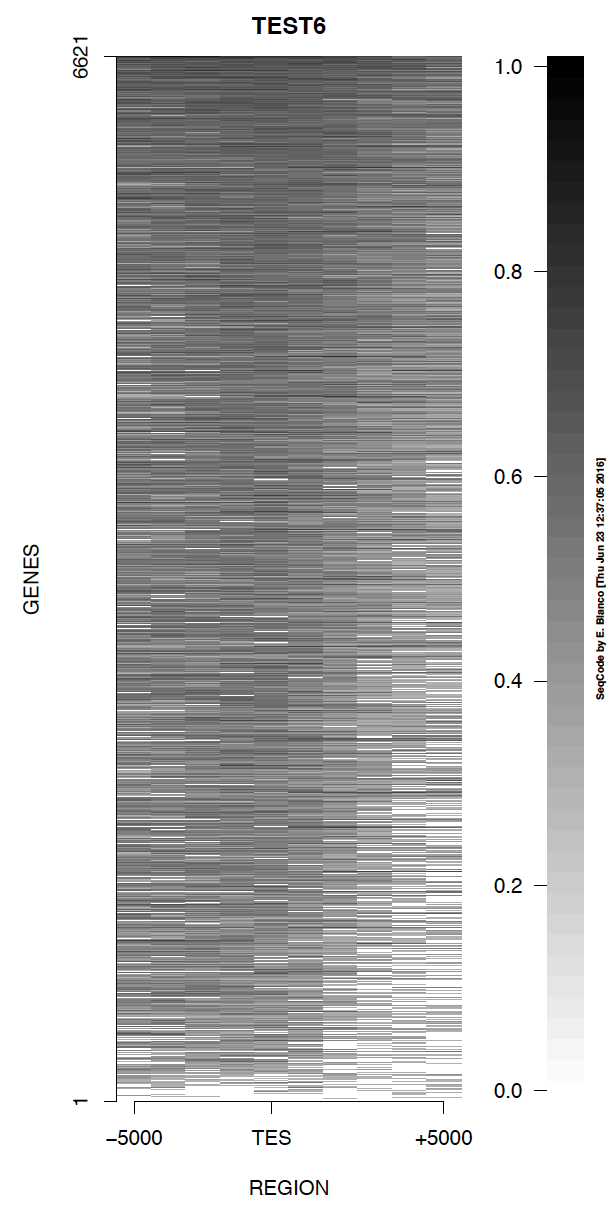
Example 6. How to decrease the resolution of the picture to generate a heat map with lower memory requirements.
> bin/produceTESmaps -v ChromInfo.txt refGene.txt SRR592584.bam H3K36me3_genes.txt TEST1 5000


Example 2. How to change the range of colors into the heat map of genes (background is white).
> bin/produceTESmaps -v -b black -f darkgoldenrod1 ChromInfo.txt refGene.txt SRR592584.bam H3K36me3_genes.txt TEST2 5000

Example 3. How to change the outer background and foreground colors of the heat map of genes.
> bin/produceTESmaps -v -B black -F darkgoldenrod1 ChromInfo.txt refGene.txt SRR592584.bam H3K36me3_genes.txt TEST3 5000


Example 4. How to change the outer background and foreground colors and the range of the heat map of genes (background is white).
> bin/produceTESmaps -v -B black -b black -F darkgoldenrod1 -f darkgoldenrod1 ChromInfo.txt refGene.txt SRR592584.bam H3K36me3_genes.txt TEST4 5000

Example 5. How to change the full schema of colors of the heat map of genes.
> bin/produceTESmaps -v -H black -B black -b black -F darkgoldenrod1 -f darkgoldenrod1 ChromInfo.txt refGene.txt SRR592584.bam H3K36me3_genes.txt TEST5 5000

Example 6. How to decrease the resolution of the picture to generate a heat map with lower memory requirements.
> bin/produceTESmaps -v -w 1000 ChromInfo.txt refGene.txt SRR592584.bam H3K36me3_genes.txt TEST6 5000

ChIPseq and RNAseq samples
Please, follow the link below for further information on how to get and preprocess the raw data of
published ChIPseq and RNAseq samples utilized in this glossary of SeqCode functions.
[SAMPLES]
[SAMPLES]