SeqCode: matchpeaksgenes
General description
Match peaks and target genes depending on the location of both sets of coordinates.
> bin/matchpeaksgenes -h SeqCode_v1.0 User commands matchpeaksgenes NAME matchpeaksgenes - a program to match peaks to genes. SYNOPSIS matchpeaksgenes [-udUDse <value>][-v][-h] <peaks.bed> <refGene.txt> OUTPUT List of matches (STDOUT). Each line is a peak associated to a transcript. OPTIONS -u : Use X bp upstream of the TSS, including gene body. -d : Use X bp downstream of the TES, including gene body. -U : Use X bp upstream of the TSS, excluding gene body. -D : Use X bp downstream of the TES, excluding gene body. -s : Use only +/- X bp around the TSS. -e : Use only +/- X bp around the TES. -v : Verbose. Display info messages. -h : Show this help. SEE ALSO SeqCode homepage: http://ldicrocelab.crg.es GitHub source code: https://github.com/eblancoga/seqcode AUTHORS Written by Enrique Blanco. SeqCode_v1.0 User commands matchpeaksgenes
Examples
Example 1. Matching peaks within the promoter (2.5 Kbp upstream of TSS) and the body of mouse genes.
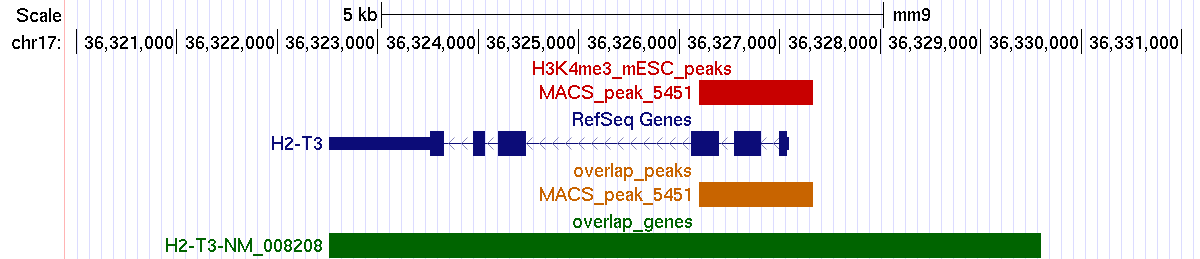
Example 2. Matching peaks within the promoter (excluding the body of target genes).
Example 3. Matching peaks within +/- 500 bps around the TSS.
> # Full output: peaks and genes
> bin/matchpeaksgenes -v -u 2500 H3K4me3_peaks.bed refGene.txt > H3K4me3_RefSeq_output.txt
> # List of target genes
> bin/matchpeaksgenes -v -u 2500 H3K4me3_peaks.bed refGene.txt | gawk '{print $5}' | sort | uniq > H3K4me3_RefSeq_target-genes.txt
> # Custom tracks for UCSC genome browser
> gawk 'BEGIN{OFS="\t"; print "track name=overlap_peaks color=200,100,0";}{print $1,$2,$3,$4}' H3K4me3_RefSeq_output.txt > H3K4me3_RefSeq_PEAKS.txt
> gawk 'BEGIN{OFS="\t"; print "track name=overlap_genes color=0,100,0";}{print $7,$9,$10,$5"-"$6}' H3K4me3_RefSeq_output.txt > H3K4me3_RefSeq_GENES.txt
UCSC genome browser screenshot:

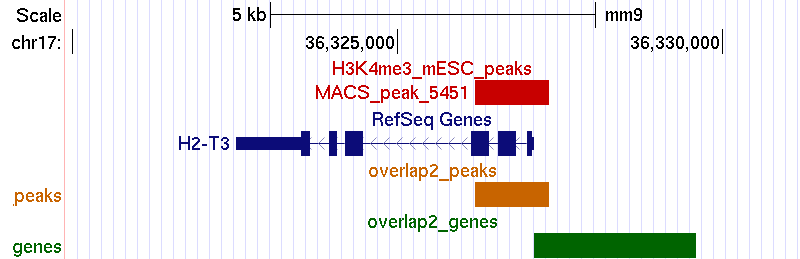
Example 2. Matching peaks within the promoter (excluding the body of target genes).
bin/matchpeaksgenes -v -U 2500 H3K4me3_peaks.bed refGene.txt > H3K4me3_RefSeq_2_output.txtUCSC genome browser screenshot:

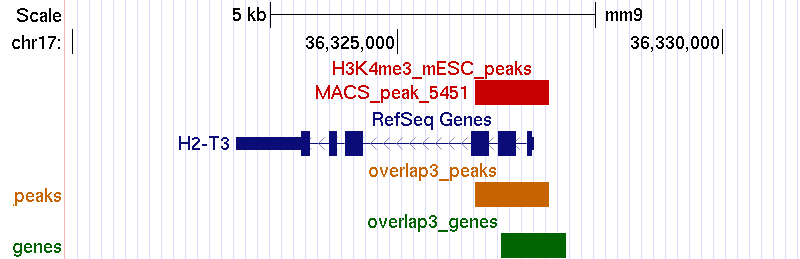
Example 3. Matching peaks within +/- 500 bps around the TSS.
bin/matchpeaksgenes -v -s 500 H3K4me3_peaks.bed refGene.txt > H3K4me3_RefSeq_3_output.txtUCSC genome browser screenshot:

ChIPseq and RNAseq samples
Please, follow the link below for further information on how to get and preprocess the raw data of
published ChIPseq and RNAseq samples utilized in this glossary of SeqCode functions.
[SAMPLES]
[SAMPLES]