SeqCode: matchpeaks
General description
Calculation of the overlap between peaks of two different ChIPseq samples.
> bin/matchpeaks -h SeqCode_v1.0 User commands matchpeaks NAME matchpeaks - a program to compare 2 lists of peaks/regions. SYNOPSIS matchpeaks [-l <bp>][-v][-x prefix][-h] <peaksA.bed> <peaksB.bed> <nameA> <nameB> OUTPUT One folder with 3 files (BED): common in both, only in A, only in B. OPTIONS -l : Minimum overlap size (default: 1). -v : Verbose. Display info messages. -x : Prefix for the output folder. -h : Show this help. SEE ALSO SeqCode homepage: http://ldicrocelab.crg.es GitHub source code: https://github.com/eblancoga/seqcode AUTHORS Written by Enrique Blanco. SeqCode_v1.0 User commands matchpeaks
Examples
Example 1. Matching two sets of peaks with all the classes of overlap.
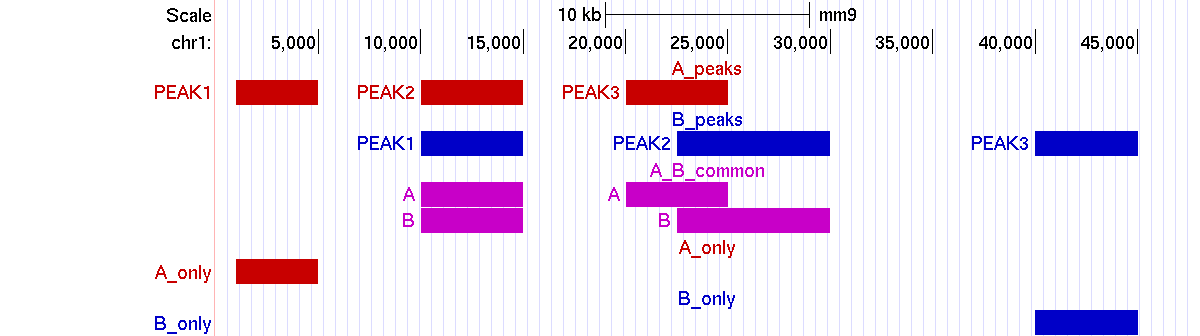
> bin/matchpeaks -v A_peaks.bed B_peaks.bed A B Peaks intersected in A: 2/3 (66.67%) Peaks intersected in B: 2/3 (66.67%) Peaks non-intersected in A: 1/3 (33.33%) Peaks non intersected in B: 1/3 (33.33%)UCSC genome browser screenshot:

ChIPseq and RNAseq samples
Please, follow the link below for further information on how to get and preprocess the raw data of
published ChIPseq and RNAseq samples utilized in this glossary of SeqCode functions.
[SAMPLES]
[SAMPLES]